核酸酶靶向切割和釋放 (CUT&RUN)是建立在免疫探測(cè)技術(shù)上的染色質(zhì)圖譜分析方法�����。在CUT&RUN中�,融合蛋白AG-微球菌核酸酶(pAG-MNase),選擇性原位切割抗體結(jié)合的染色質(zhì)(Figure 1)�。該方法與二代測(cè)序(NGS)兼容����,可提供組蛋白翻譯后修飾(PTMs)和染色質(zhì)相關(guān)蛋白(如轉(zhuǎn)錄因子[TFs])的高分辨率全基因組圖譜��。
FIGURE 1?Overview of the CUTANA? CUT&RUN protocol.
產(chǎn)品優(yōu)勢(shì)
?CUTANATM?Library Prep Kit 是第一個(gè)專門為CUT&RUN分析開發(fā)的文庫構(gòu)建試劑盒�����。
?對(duì)CUT&RUN的方法進(jìn)行了優(yōu)化�����,對(duì)比多用途或ChIP-seq文庫構(gòu)建試劑盒更具優(yōu)勢(shì)��。
?對(duì)于CUT&RUN產(chǎn)生的有限輸入���,工作流程非常穩(wěn)定�,為使用0.5-10?ng DNA的Illumina??NGS提供了高質(zhì)量的文庫����。
?試劑盒包含CUT&RUN文庫制備所需的所有材料(酶、引物�����、DNA純化磁珠、緩沖液和PCR管)��。
?可輕松搭配CUTANA? CUT&RUN試劑盒(EpiCypher 14-1048)或CUT&RUN Protocol (EpiCypher.com/protocols)使用�����,實(shí)現(xiàn)工作流程集成����,高通量檢測(cè)和保證結(jié)果的可靠性���,降低實(shí)驗(yàn)成本�。
?可以有效制備組蛋白PTMs和染色質(zhì)相關(guān)蛋白(如TFs)CUT&RUN DNA的文庫���。
?
Multiplexing Primers
Primer Set 1 includes i5 primers 1-8 and i7 primers 1-6?
Primer Set 1 includes i5 primers 1-8 and i7 primers 7-12
保存條件
OPEN KIT IMMEDIATELY and store components at room temperature and -20°C as indicated (see Kit Manual for full instructions). Stable for 6 months upon date of receipt.
Room Temperature (RT) | -20℃ |
8-strip Tubes | End Prep Enzyme | Ligation Enhancer |
SPRIselectreagent manufactured by Beckman Coulter, Inc. | End Prep Buffer | High Fidelity 2X PCR Master Mix |
0.1X TE Buffer | Adapter for Illumina? | U-Excision Enzyme |
| Ligation Mix | i5 and i7 Primers |
數(shù)據(jù)示例
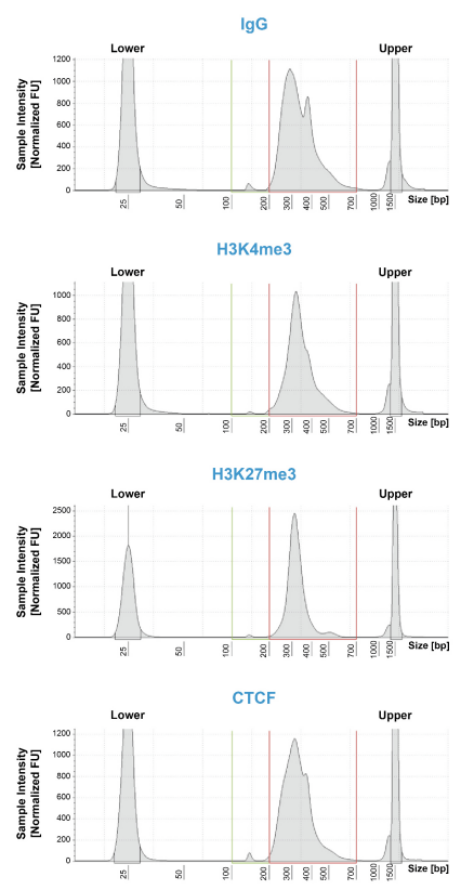
|
FIGURE 1: CUT&RUN DNA Fragment Size Distribution Analysis. CUT&RUN was ?performed as described above.?Library DNA was ?analyzed by Agilent TapeStation? , which ?confirmed that mononucleosomes were??predominantly enriched in CUT&RUN (~300 bp ?peaks represent 150 bp nucleosomes + ?sequencing?adapters). Peaks at ~380 bp ?correspond to the SNAP-CUTANA??K-MetStat Panel of spike-in controls(EpiCypher 19-1002). |
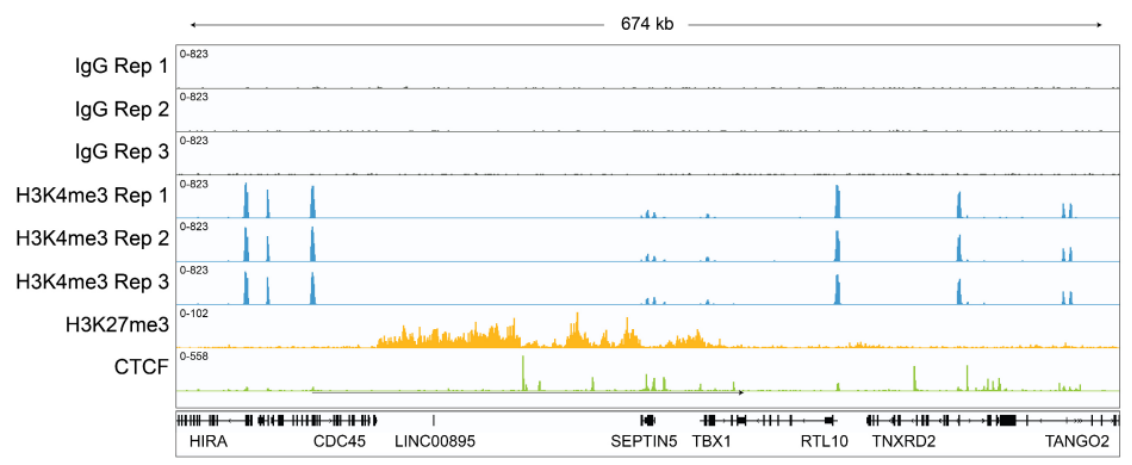
|
FIGURE 2: Representative Gene Browser Tracks. CUT&RUN was performed as described above. A representative?674 kb window at the SEPTIN5 gene is shown for three replicates (”Rep”) of IgG and H3K4me3 antibodies,?as well as individual tracks for H3K27me3 and the transcription factor CTCF, demonstrating the robustness?and reproducibility of the workflow with a variety of targets. Sequencing libraries prepared with the CUTANA?CUT&RUN Library Prep kit produced the expected genomic distribution for each target. Images were generated?using the Integrative Genomics Viewer (IGV, Broad Institute) |
訂購(gòu)詳情
貨號(hào) | 產(chǎn)品名稱 | 規(guī)格 |
14-1001 | CUTANA? CUT&RUN Library Prep Kit?with Primer Set 1 | 48 Reactions |
14-1002 | CUTANA? CUT&RUN Library Prep Kit?with Primer Set 2 | 48 Reactions |
?

如需了解更多詳細(xì)信息或相關(guān)產(chǎn)品��,請(qǐng)聯(lián)系EpiCypher中國(guó)授權(quán)代理商-欣博盛生物?
全國(guó)服務(wù)熱線: 4006-800-892 ? ? ??郵箱: market@neobioscience.com?
深圳: 0755-26755892 ? ? ? ??北京: 010-88594029 ???????????
廣州:020-87615159????????? ?上海: 021-34613729
代理品牌網(wǎng)站: www.yuebanme.com?
自主品牌網(wǎng)站: www.neobioscience.net